Full list
- VanBuren R*, Wai CM, Pardo J, Yocca AE, Wang X, Wang H, Edger PP, Bennetzen JL, Michael T. (2020) Subgenome dynamics and stability in the allotetraploid grain crop tef. Nature Commications, BioRxiv 58712
- Baetsen-Young A, Wai CM, VanBuren R, Day B. Fusarium virguliforme Transcriptional Plasticity Is Revealed by Host Colonization of Corn vs. Soybean. (2019) Plant Cell, https://doi.org/10.1105/tpc.19.00697
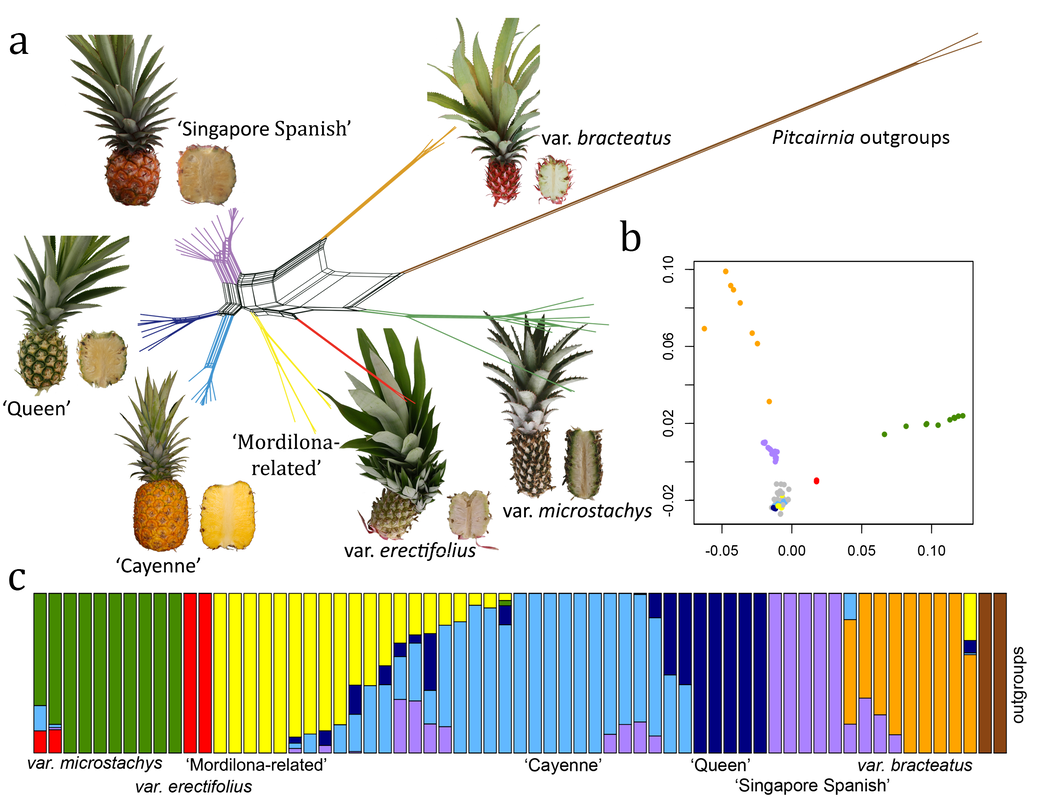
- Chen L-Y#, VanBuren R#, Paris M#, Zhou H, Zhang X, Wai CM, Yan H, Chen S, Alonge M, Ramakrishnan S, et al. (2019) The bracteatus pineapple genome and domestication of clonally propagated crops. Nature Genetics doi: 10.1038/s41588-019-0506-8
- Azodi CB, Pardo J, VanBuren R, de los Campos G, Shiu SH. (2020) Transcriptome-based prediction of complex traits in maize. Plant Cell, https://doi.org/10.1105/tpc.19.00332
- Edger PP, Poorten TJ, VanBuren R, et al. Origin and evolution of the octoploid strawberry genome. Nature genetics 2019
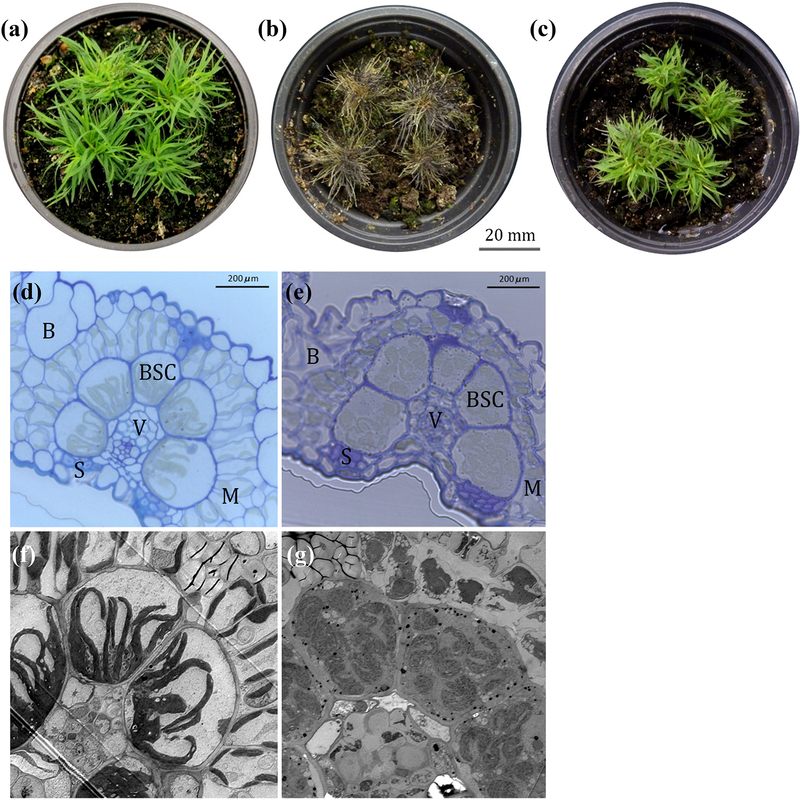
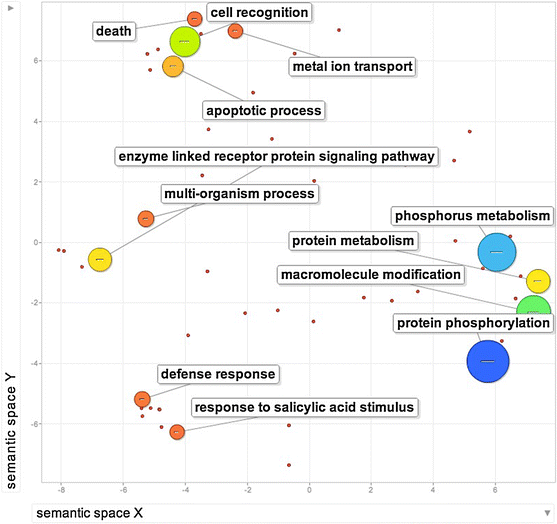
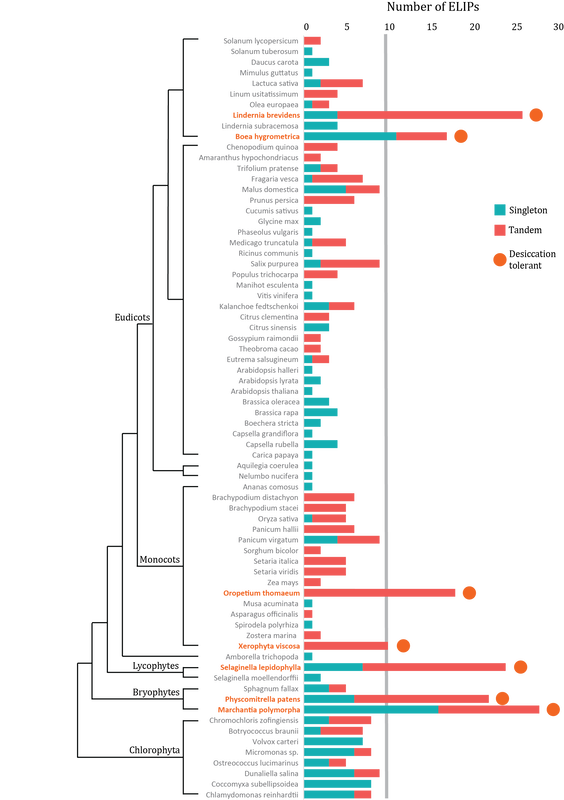
- VanBuren R*, Pardo J, Wai CM, Evans S, Bartels D. Massive tandem proliferation of ELIPs supports convergent evolution of desiccation tolerance across land plants. Plant physiology 2019, https://doi.org/10.1104/pp.18.01420.
- Colle M, et al. VanBuren R, Jiang N, Edger PP. Subgenome dominance and evolution of phytonutrient pathways in allopolyploid blueberry. GigaScience 2019, https://doi.org/10.1093/gigascience/giz012
- Mower J, Ma PF, Grewe F, Taylor A, Michael TP, VanBuren R, Yin-Long Q. Lycophyte plastid genomics: extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New phytologist 2018, https://doi.org/10.1111/nph.15650
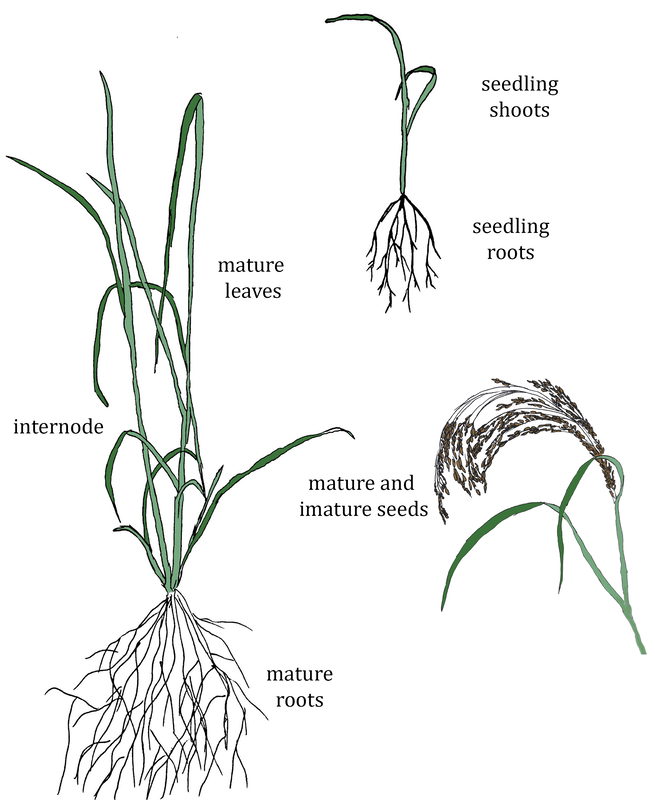
- VanBuren R*, Wai CM, Keilwagen J, Pard J. A chromosome scale assembly of the model desiccation tolerant grass Oropetium thomaeum. Plant direct 2018.
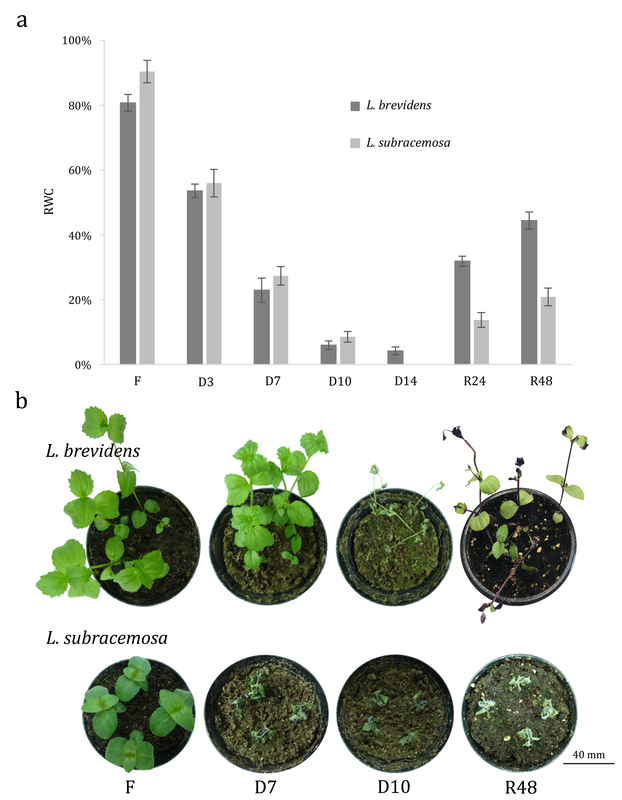
- VanBuren R*, Wai CM, Pardo J, Giarola V, Ambrosini S, Somg X, Bartels D. Desiccation tolerance evolved through gene duplication and network rewiring in Lindernia. Plant cell 2018, DOI: https://doi.org/10.1105/tpc.18.00517. *corresponding author
- Zhang J… (114 coauthors), VanBuren R, Paterson AH, Nagai C, Ming R. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nature genetics 2018, 50:1565–1573.
- VanBuren R*, Wai CM, Colle M, Wang J, Sullivan S, Bushakra JM, Liachko I, Vining KJ, Dossett M, Finn CE, Jibran R. A near complete, chromosome-scale assembly of the black raspberry (Rubus occidentalis) genome. GigaScience 2018, 7(8):giy094.
- Bushakra JM, Dossett M, Carter KA, Vining KJ, Lee JC, Bryant DW, VanBuren R, Lee J, Mockler TC, Finn CE, Bassil NV. Characterization of aphid resistance loci in black raspberry (Rubus occidentalis L.). Molecular breeding 2018, 38(7):83.
- Bird KA, VanBuren R, Puzey JR, Edger PP. The causes and consequences of subgenome dominance in hybrids and recent polyploids. New phytologist 2018
- Edger PP, McKain MR, Bird KA, VanBuren R. Subgenome assignment in allopolyploids: challenges and future directions. Current opinion in plant biology. 2018 42:76-80.
- Jibran R, Dzierzon H, Bassil N, Bushakra, JM, Edger P, Sullivan S, Finn CE, Dossett M, Vining, KJ, VanBuren R, and Mockler TC, 2018. Chromosome-scale scaffolding of the black raspberry (Rubus occidentalis L.) genome based on chromatin interaction data. Horticulture research, 5(1), p.8.
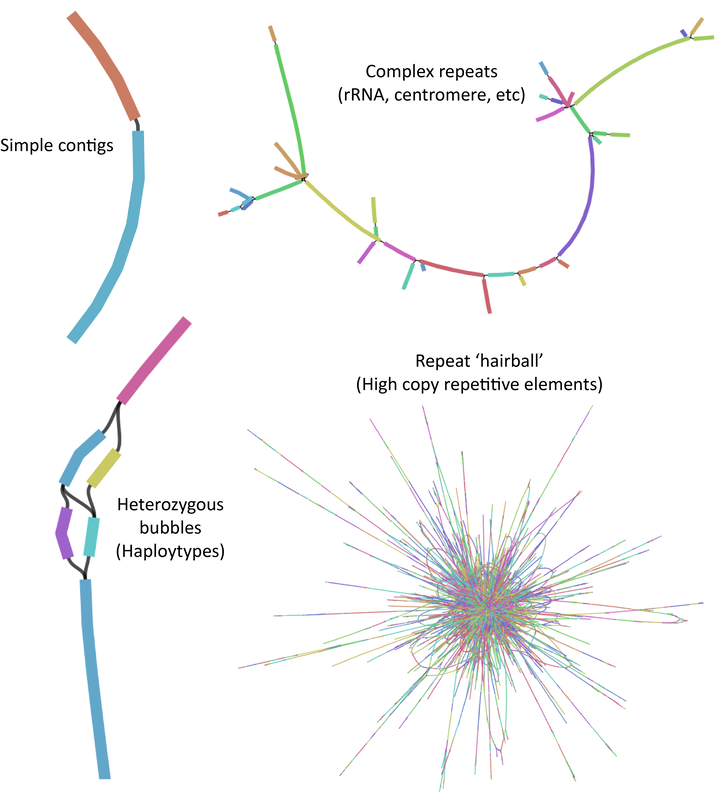
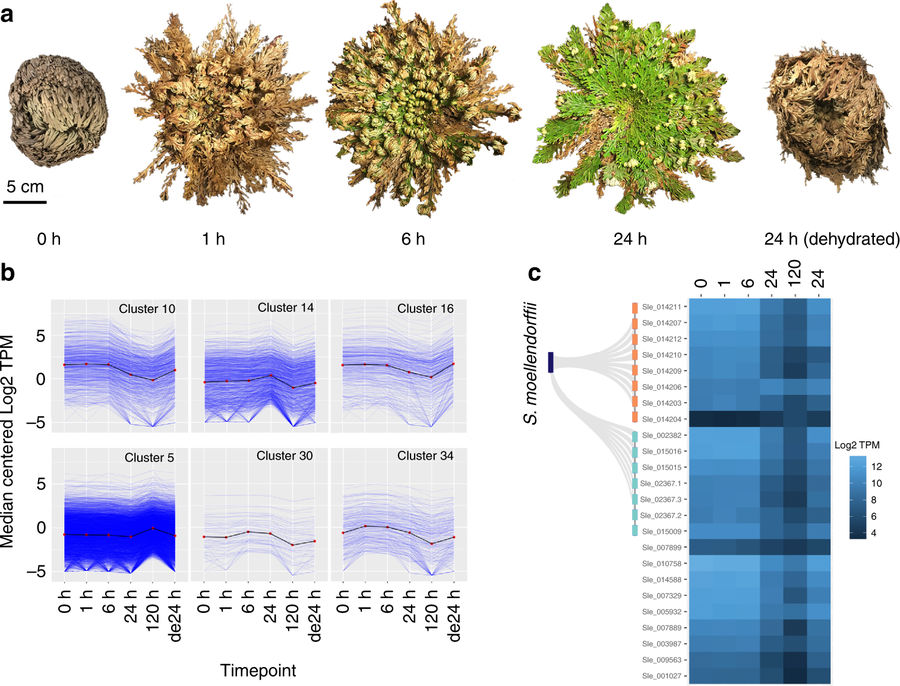
- VanBuren R*, Wai CM, Ou S, Pardo J, Bryant D, Jiang N, Mockler TC, Edger P, Michael T. Extreme haplotype variation in the desiccation-tolerant clubmoss Selaginella lepidophylla. Nature communications 2018, 9:13. *corresponding author
- Jibran R, Dzierzon H, Bassil N, Bushakra JM, Edger PP, Sullivan S, Finn CE, Dossett M, Vining KJ, VanBuren R: Chromosome-scale scaffolding of the black raspberry (Rubus occidentalis L.) genome based on chromatin interaction data. Horticulture research 2018, 5:8.
- VanBuren R*, Wai J, Zhang Q, Song X, Edger PP, Bryant D, Michael TP, Mockler TC, Bartels D: Seed desiccation mechanisms co‐opted for vegetative desiccation in the resurrection grass Oropetium thomeaum. Plant, cell & environment 2017.
- VanBuren R: Desiccation tolerance: Seedy origins of resurrection. Nature plants 2017, 3:17046.
- Yang X, Hu R, … VanBuren R, (33 out of 50 authors) et al. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. 2018 Nature communications. 8: 1899.
- Edger PP*, VanBuren R*, Colle M, Poorten TJ, Wai CM, Niederhuth CE, Alger EI, Ou S, Acharya CB, Wang J: Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. GigaScience 2017.
- Wai CM*, VanBuren R*, Zhang J, Huang L, Miao W, Edger PP, Yim WC, Priest H, Meyers BC, Mockler TC, Smith AJ, Cushman JC, Ming R. 2017 Temporal and spatial transcriptomic and microRNA dynamics of CAM photosynthesis in pineapple. The Plant Journal. 10.1111/tpj.13630
- VanBuren R. 2017 Desiccation tolerance: Seedy origins of resurrection. Nature Plants. 3; 17046.
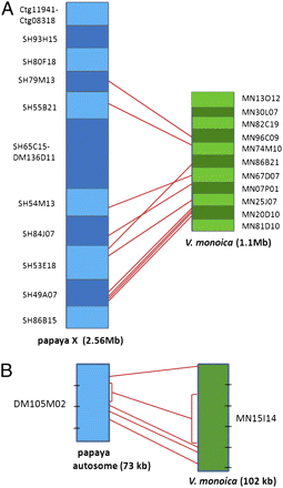
- VanBuren R, Wai CM, Zhang J, Han J, Arro J, Lin Z, Liao Z, Yu Q, Wang ML, Zee F, Moore RC, Charlesworth D, Ming R. 2016 Extremely low nucleotide diversity in the X-linked region of papaya caused by a strong selective sweep. Genome Biology. Nov 28;17(1):230.
- VanBuren R, Bryant R, et al. 2015 The genome of Black Raspberry (Rubus occidentalis). The Plant Journal. DOI: 10.1111/tpj.13215.
- Ming R*, VanBuren R*, Wai CM*, Tang H* et al. 2015 The pineapple genome and the evolution of CAM photosynthesis. Nature Genetics. DOI: 10.1038/ng.3435. *equal contribution
- VanBuren R, Bryant D, Edger PP, Tang H, Burgess D, Challabathula D, Spittle K, Hall R, Gu J, Lyons E, Freeling M, Bartes D, Hallers BT, Hastie A, Michael TP, Mockler TC. 2015 Single-molecule sequencing of the desiccation tolerant grass Oropetium thomaeum. Nature 527, 508-511.
- Zhang Q, Liu C, Liu Y VanBuren R, Yoa X, Zhong C, Huang H 2015 High density interspecific genetic maps of kiwifruit and the identification of sex specific markers. DNA Research dsv019.
- Bushakra J, Bryant D, Dossett M, Vining K, VanBuren R, Gilmore B, Lee J, Mockler TC, Finn C, Bassil N. 2015 A genetic linkage map of black raspberry (Rubus occidentalis) and the mapping of Ag 4 conferring resistance to the aphid Amphorophora agathonica. Theoretical and Applied Genetics 128,1631-1646.
- Michael TP, VanBuren R*. 2015 Progress, challenges and the future of crop genomics Current opinion in plant biology 24, 71-81. *Corresponding author
- VanBuren R, Zeng F, Chen C, Zhang J, Wai CM, Han J, Aryal R, Gschwend AR, Wang J, Na JK, Huang L, Zhang L, Miao W, Gou J, Arro J, Guyot R, Moore RC, Wang M, Zee F, Charlesworth D, Moore PH, Yu Q, Ming R. 2015 Domestication of Yh chromosome in papaya. Genome Research 25, 524-533.
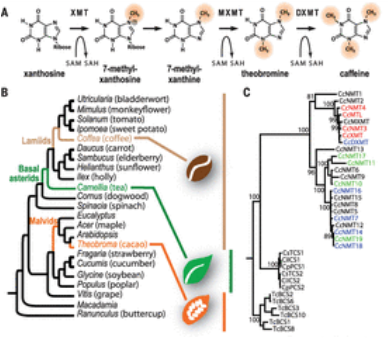
- Deneud F … VanBuren R (48 out of 62 alphabetical authors) et al. 2014 The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345, 1181-118
- Zhang Q, Li L, VanBuren R, Liu Y, Yang M, Han Y, Ming R. 2014 Optimizing linkage mapping strategy using genome sequence based markers and construction of high-density sacred Lotus genetic maps. BMC Genomics 15, 372.
- Lum G, VanBuren R, Ming R, Min XJ. 2013 Secretome Prediction and Analysis in Sacred Lotus (Nelumbo nucifera Gaertn.) Tropical Plant Biology 6, 131-137.
- VanBuren R, Walters B, Jia Min X, Ming R. 2013 Analysis of expressed sequence tags and alternative splicing genes in sacred lotus ('Nelumbo nucifera'Gaertn.) Plant Omics. 6, 311.
- Ming, R*, VanBuren R*, Liu Y, Yang M, et al. (84 coauthors) 2013 The genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.) Genome Biology 14, R41. * equal contribution
- VanBuren R, Ming R. 2013 Chloroplast DNA accumulation in the recently evolved Y chromosome of Carica papaya. Molecular Genetics and Genomics 288, 277-284.
- VanBuren R, Ming R. 2013 Dynamic transposable element accumulation in the nascent sex chromosomes of papaya. Mobile Genetic Elements 3,1-5.
- Yang M, Han Y, VanBuren R, Ming R, Xu L, Han Y, Liu Y. 2012 Genetic linkage maps for Asian and American lotus constructed using novel SSP markers derived from the genome of sequenced cultivar. BMC genomics 13:653.
- Wang J, Na J, Yu Q, Gschwend A, Han J, Zeng F, Aryal, VanBuren R, et al. 2012 Dynamic rearrangements, genetic degeneration, and gene trafficking in the papaya nascent Yh chromosome. PNAS 109,13710-13715.
- Gschwend A, Yu Q, Zeng F, Han J, VanBuren R Charlseworth D, Moore P, Paterson A, Ming R. 2012 Rapid divergence and expansion of the X chromosome in papaya. PNAS 109, 13716-13721.
- VanBuren R, Li J, Zee F, Zhu J, Liu C, Arumaganathan A, Ming R. 2011 Longli is not a hybrid of longan and lychee as revealed by genome size analysis and trichome morphology. Tropical Plant Biology. 4,228-236.
- Hudson AO, Ahmad N, VanBuren R, Savka MA. 2010 Sugarcane and Grapevine Endophytic Bacteria: Isolation, Detection of quorum Sensing signals and Identification by 16S v3 rDNA Sequence analysis. Current Research Technology and Education Topics in Applied Microbiology and Microbial Biotechnology 13,801-806.